In July 2021, a Google project called DeepMind published AI-enabled predictions of nearly every protein in the human genome. This was described in various articles by evolutionary and computational biologists as a “big deal,” “game changer”, and “paradigm shift”.1 We agree. Here’s why:
There are ~20,000 proteins in the human proteome. Each one is constantly cycling through dynamic conformations (shapes) and forming complex protein-protein interactions (governed by laws of physics) that ultimately drive biological function. Drugs work by interacting with proteins to alter their conformation, signaling capacity and/or ability to bind other proteins and, in doing so, alter their function. A natural starting point for any hypothesis-driven drug development campaign is a basic understanding of a protein that is known to cause a particular disease. Examples include, a mutant protein in cancer that is stuck in the “grow” conformation or a mutant protein in a rare disease whose conformation can’t be metabolized appropriately and therefore accumulates maliciously within cells. However, knowing exactly what a particular protein looks like so that you can bind it with a novel drug is actually extremely difficult.
Proteins are: very small; hard to isolate and purify; constantly in motion; highly homologous (having a similar structure). To properly characterize them at sufficiently clear enough resolution, highly specialized researchers have developed laborious and technical strategies to isolate protein crystals (something Manny Rocha, a Driehaus analyst who completed his PhD in biology at the University of Chicago, described as “black magic”) and then bombard them with electrons to produce microscopic outlines. When those outlines are taken from multiple different angles and brought together, they form a picture of a 3D shape. This technique is called x-ray crystallography. The process is considered so difficult that “solving” a protein’s structure (that is, determining its 3D shape) alone can be sufficient for publication in august journals, and that as a short-cut many researchers only try to solve a smaller portion of a protein at a time.
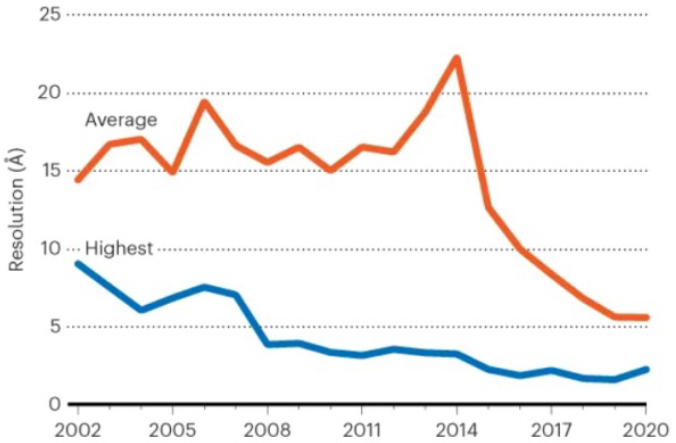
In 2003, there were fewer than 10 solved protein and protein fragment structures released by the Electron Microscopy Data Bank. By 2019, that number increased to more than 13,000 and was growing by more than 3,000 per year (see Exhibit 5 below). Excitingly, not only is the number of solved structures increasing, but the resolution of these structures is also improving with technological advancements, going from an average of 15-20 angstroms2 from 2002-2014 to <10 after 2015 (see Exhibit 6 below).
Exhibit 1: Electron Microscopy Data Bank Total Number of Structures Released Annually, 2002-2021 YTD3
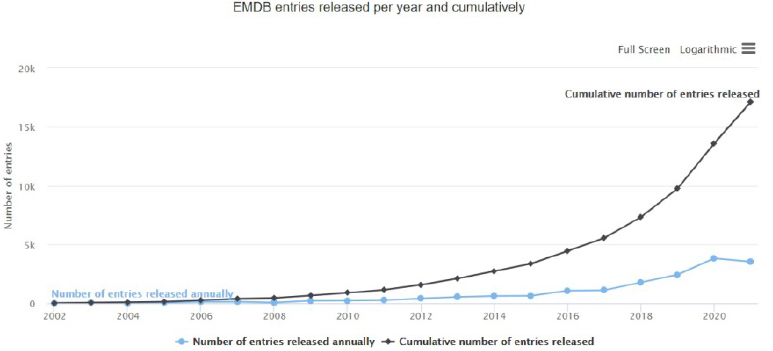
Exhibit 2: Electron Microscopy Data Bank Resolution of Structures Released Annually (Measured in Angstroms), 2002-20204
More than 13,000 proteins and protein fragments may sound like a majority of the 20,000 proteins in the human proteome, but recall the following: i) the database includes protein fragments in addition to full proteins; and ii) any given protein can have a vast number of different stable and intermediate conformations or other post-translational modifications, all of which can drive significant structural distinctions. As a result, only about 1/3 of the proteins in the proteome are considered to have “solved” full-size structures, and even those have likely only a fraction of the total potential conformations fully elucidated. Mutated proteins add substantially to the “unsolved” category.
With DeepMind’s published predictions of nearly all of the proteins in the proteome, the number of “solved” full-size structures took a great leap forward. As a result, one of the key bottlenecks to generating novel hypothesis-driven medicine – the foundation of Driehaus Life Sciences’ largest investments – has been dramatically loosened, such that the realm of possibilities for future rationally designed medicines just expanded exponentially. To help contextualize the impact of this, consider the opinion of one evolutionary biologist from the Max Planck Institute in Germany, who had been working to solve the structure of a bacterial protein: “The model from [Deepmind] gave us our structure in half an hour, after we had spent a decade trying everything.”5
Before we get too caught up in the excitement, it’s important to note that DeepMind has limitations. First, the predictions are just that: predictions based on previously solved structures. Until they’ve been confirmed experimentally, it will be difficult to know how much validity to ascribe to them. Second, many of the predictions from DeepMind appear maddeningly incomplete. Comparing DeepMind’s predictions of a key target protein to proprietary structural information gleaned over years of experimental research, the Head of R&D at a multi-billion dollar company with whom we have a strong relationship told us DeepMind’s prediction “looks like a bowl of spaghetti.”6 Rather than replacing experimental methods, this expert believes DeepMind will serve as a useful starting point to speed up experimental solutions. Even then, though, there will still be the limitation of a single, static conformation, versus the reality of proteins, which cycle through stable and intermediate conformations.
It’s tempting to believe that DeepMind’s predictions will be leverageable by AI-enabled technology companies to speed up drug development and make it more predictive systematically. Undoubtedly, many will try. However, we don’t believe that this breakthrough on its own will yield high probability-of-success development-stage drugs. Instead, we believe it will be a necessary but insufficient precondition for success that will still require the right people, strategy, execution, and financial partners. To quote some of the savviest drug developers we’ve met over the past decade-plus:
- “Discovering high quality selective compounds is not easy and there are no real platform-based shortcuts – work can take many years."
- "Structure-based drug design is a ‘must have,’ but the tools have become commoditized – program differentiation, speed, and probability of success are determined by the people operating the tools."
- "Each product is a unique challenge – success comes from talented people working together in an environment that provides the time, resources and intellectual honesty to develop great medicines.”7
For companies doing this, we intend to be an excellent financial and strategic partner – in private and public markets – and in doing so achieve the objectives of the fund and the betterment of mankind.
1Callaway, “DeepMind’s AI predicts structures for a vast trove of proteins,” Nature, July 22, 2021. Accessed 10/22/2021 from here. Also Callaway, “‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures,” Nature, November 30, 2020. Accessed 10/22/2021 from here.
2An angstrom is a unit of measurement equal to one ten millionth of a millimeter. It is a unit of measurement commonly used in molecular biology.
3Electron Microscopy Data Bank, “a public repository for electron cryo-microscopy volume maps and tomograms of macromolecular complexes and subcellular structures. It covers a variety of techniques, including single-particle analysis, electron tomography, and electron crystallography.” Accessed 10/22/2021 from here.
4Callaway, “Revolutionary cryo-EM is taking over structural biology,” Nature, February 10, 2020. Accessed 10/24/2021 from here.
5Callaway, “‘It will change everything’: DeepMind’s AI makes gigantic leap in solving protein structures,” Nature, November 30, 2020. Accessed 10/24/2021 from here.
6Driehaus interview, July 2021. Source anonymized as we have not sought his permission to quote him.
7Hyman, “Discovery of mutant selective PI3kalpha and isoform-selective FGFR3 inhibitors – Insights from the Loxo Oncology at Lilly Discovery Model,” Molecular Targets and Cancer Therapeutics Conference: October 7-10.
This information is not intended to provide investment advice. Nothing herein should be construed as a solicitation, recommendation or an offer to buy, sell or hold any securities, market sectors, other investments or to adopt any investment strategy or strategies. You should assess your own investment needs based on your individual financial circumstances and investment objectives. This material is not intended to be relied upon as a forecast or research. The opinions expressed are those of Driehaus Capital Management LLC (“Driehaus”) as of November 2021 and are subject to change at any time due to changes in market or economic conditions. The information has not been updated since November 2021 and may not reflect recent market activity. The information and opinions contained in this material are derived from proprietary and non-proprietary sources deemed by Driehaus to be reliable and are not necessarily all inclusive. Driehaus does not guarantee the accuracy or completeness of this information. There is no guarantee that any forecasts made will come to pass. Reliance upon information in this material is at the sole discretion of the reader.
Other Commentaries
Data Center
By Ben Olien, CFA
Driehaus Micro Cap Growth Strategy March 2024 Commentary with Attribution
By US Growth Equities Team